DNA origami showcases "mechanical frustration"
DNA origami showcases "mechanical frustration"
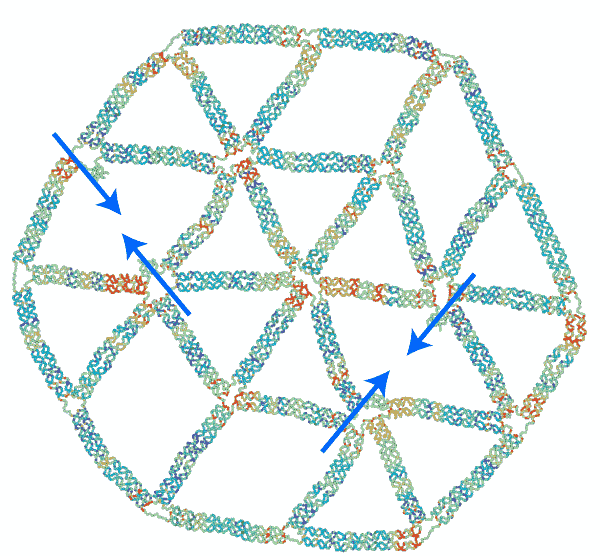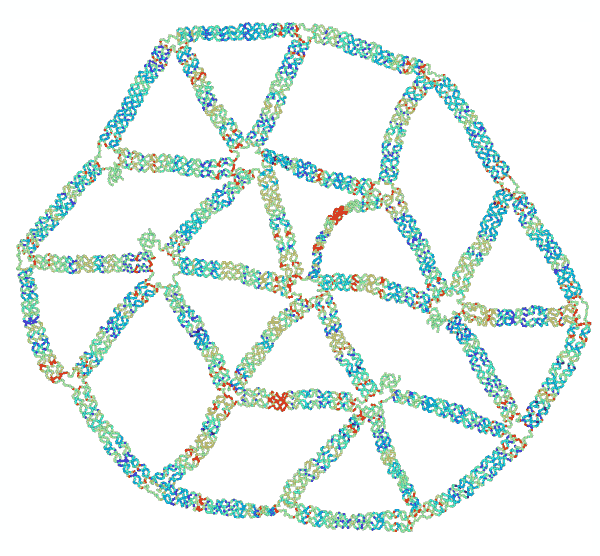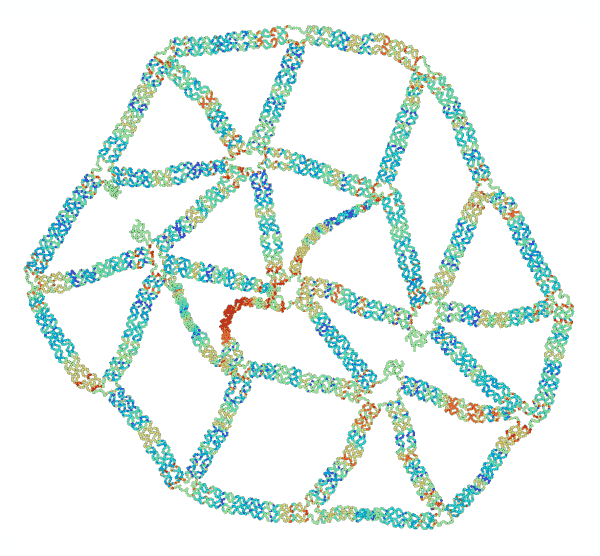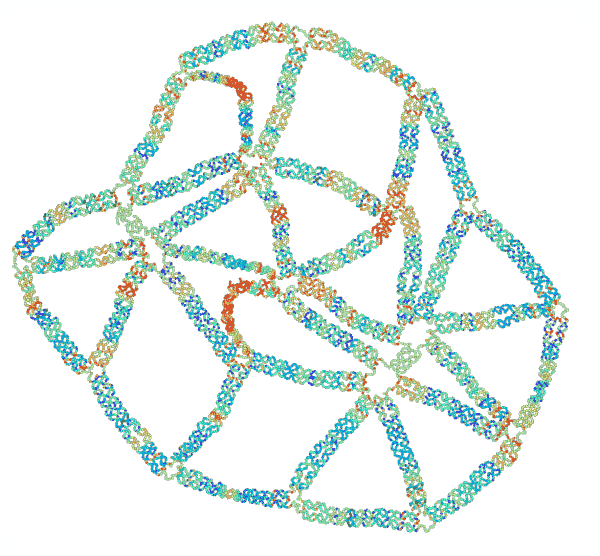
“The concept of ‘frustration’ is well known in the study of magnetism,” said Jong Hyun Choi, professor of mechanical engineering. “Two particles have opposing charges, and they point 180 degrees opposite to each other. But when you have three particles in a triangle, they can’t all point away from each other, and that imbalance is called ‘frustration.’ We are duplicating that concept, but with mechanical forces instead of magnetic.”
In Choi’s lab, they use CAD software to assemble nanoscale mechanical structures using DNA strands as building blocks. Because the four basic components of DNA (A, C, G, and T) always connect the same way, their CAD designs can be translated into chemical DNA sequences, causing the strands to self-assemble into any shape they want.
To test their theories of mechanical frustration, they first formed the simplest 2D shape possible: a triangle. Connecting several equilateral triangles, they then formed a lattice structure. Next, they experimented with removing specific strands to induce frustration.
“We found that removing certain strands created adaptable and inadaptable structures,” said Anirudh Madhvacharyula, Ph.D. student in Choi’s lab and lead author of the paper. “If you remove one set of strands, the structure collapses in an adaptable way, meaning the stress is distributed evenly. But if you remove another set, it becomes an inadaptable (frustrated) structure, where all of the unbalanced mechanical strain is localized to one part of the structure. That’s mechanical frustration.”
Their research has been published in Nature Communications.
“The end goal of this work is to build customizable nanoscale structures, with a tailored mechanical response,” said Choi. “Using DNA is a great stepping stone to that. If we were to 3D print this on a human scale, the design is already set and we couldn’t actuate it without expending a lot of energy. But with DNA strands, we can add or remove certain strands using very little energy, which causes the structure to switch between two states exactly as we predicted.”
“We first design and simulate the structure in the computer,” said Madhvacharyula. “That shows us where the mechanical strain will occur, just like an engineer using finite element analysis to design and evaluate a human-scale object. Then we run the experiment and use atomic force microscopy to witness the results, because these DNA structures are less than 100 nanometers across. We tune the design further using multiple rounds of simulations and experiments to demonstrate a variety of states based on the strands removed. It has become a powerful computational design tool.”
So what are the real-world applications of mechanical frustration? While many nanoscale experiments focus on biomedical or nanomanufacturing outcomes, Choi has a more ambitious goal.
“We envision this forming the basis of new nanoscale mechanical computing systems,” he said. “Typical computers use a binary electrical state: 1 or 0, on or off. With the mechanical frustration we’ve shown, these predictable DNA responses could function much like those on-or-off bits. Our bodies already use DNA to store tremendous amounts of biological information, so maybe we can leverage this in the future to store digital information, or perform computations in a new way.”
“In the end, this is curiosity-driven research,” Choi said. “The possibilities are infinite.”
Source: Jong Hyun Choi, jchoi@purdue.edu; Anirudh Madhvacharyula, amadhvac@purdue.edu
Writer: Jared Pike, jaredpike@purdue.edu, 765-496-0374
Realizing mechanical frustration at the nanoscale using DNA origami
Anirudh S. Madhvacharyula, Ruixin Li, Alexander A. Swett, Yancheng Du, Seongmin Seo, Friedrich C. Simmel & Jong Hyun Choi
https://doi.org/10.1038/s41467-025-60492-z
ABSTRACT: Structural designs inspired by physical and biological systems have been previously utilized to develop mechanical metamaterials with enhanced properties based on clever geometric arrangement of constituent building blocks. Here, we use the DNA origami method to realize a nanoscale metastructure exhibiting mechanical frustration, a counterpart of the well-known phenomenon of magnetic frustration. By selectively actuating reconfigurable struts, it adopts either frustrated or non-frustrated states, each characterized by distinct free energy profiles. While the non-frustrated state distributes the strain homogeneously, the frustrated mode concentrates it at a specific location. Molecular dynamics simulations reconcile the contrasting behaviors and provide insights into underlying mechanics. We explore the design space further by tailoring responses through structural modifications. Our work combines programmable DNA self-assembly with mechanical design principles to overcome engineering limitations encountered at the macroscale to design dynamic, deformable nanostructures with potential applications in elastic energy storage, nanomechanical computation, and allosteric mechanisms in DNA-based nanomachinery.
